SOFTWARE
Many software packages exist for analyzing small angle scattering data. Whilst the list below is not comprehensive, it may help guide SAS practitioners by sorting them according to criteria, such as area of application, target operating system, etc.. Suggestions for additional entries are welcome. Please send your suggestions to info@smallangle.org. When doing so, please format your additions in a similar manner to those below before submission.
CONTENTS
Software for Data Reduction and Visualisation
Software for the Analysis of Biomolecular & Fibre Systems
Software for Computing Scattering from Coordinate Data
Software for Peak-Fitting and Correlation Function Analysis
Software for Parameter Distribution Analysis
Software for the Analysis of Grazing Incidence SAS
Software for the Analysis of Reflectometry Data
NOTES
canSAS Format: does the application read (R) or write (W) canSAS standard data formats (1D = I(Q) vs Q, nD = multi-dimensional)?
CCP13 = Collaborative Computational Project #13; a UK-funded software project - no longer in existence
|
Software for Data Reduction and Visualisation (also see Other Software) |
||||||||
|
Application |
canSAS Format |
Description |
Author |
Documentation |
Windows |
Linux |
Unix |
Mac |
|
BHplot |
no |
SAXS-oriented data reduction and display incorporating MatLab |
M Sztucki |
n/a |
||||
|
BioXTAS RAW |
no |
Open-source, python, multiplatform, GUI-based program focused on easy reduction of 2D data to 1D data and preliminary analysis of 1D data for biological SAS experiments. |
S Skou & J Hopkins |
|||||
|
BS-2D |
no |
A Windows-based version of BSL. |
N Koubassova |
n/a |
n/a |
n/a |
||
|
BSL |
no |
An antiquated line-mode application for the manipulation of 2D SAXS image data. |
J Bordas & G Mant |
n/a |
n/a |
|||
|
DAWN |
yes (write only) |
Extensible open-source Eclipse-based scientific data analysis & visulisation workbench |
Diamond/ESRF/EMBL |
Ubuntu (test only) |
n/a |
|||
|
DPDAK |
|
Open source tool for (online) analyzing large sequences of small angle scattering data. NB: Has FIT2D and GISAXS plugins. |
DESY/MPIKG |
|
|
|||
|
EDFplot |
no |
Processing of 2D SAXS images |
M Sztucki |
n/a |
||||
|
ESRF SAXS Programs |
|
General data manipulation routines |
P Boesecke |
n/a |
||||
|
FIT2D |
no |
Flexible data analysis and visualisation program popular with the SAXS community. |
A Hammersley |
n/a |
n/a |
|||
|
GRASP |
no |
MatLab script application designed for the visualisation & processing of ILL SANS data. |
C Dewhurst |
Webpage. NB: Requires MatLab. | ||||
|
IMAGEJ |
no |
Java-based image processing program. |
W Rasband / NIH |
Webpage. NB: Requires Java. | W32/W64 | Linux32/Linux64 | n/a | OSX |
|
IMAGEJ Plugins |
no |
Plugins for ImageJ for data reduction and geometrical corrections (Open Source) |
Documentation | W32/W64 | Linux32/Linux64 | Unix | OSX | |
|
INDRA |
no |
IGOR script application for the reduction of APS USAXS data. NB: Requires IGOR. |
J Ilavsky |
Webpage | Windows | Mac | ||
| NIKA | 1D W |
IGOR script application for the reduction of APS 2D data. NB: Requires IGOR. |
J Ilavsky | Webpage | Windows | Mac | ||
| 'NIST' | 1D R/W |
IGOR script application for the reduction of NIST SANS/USANS data. |
S Kline | Webpage. NB: Requires IGOR. | ||||
|
Mantid |
1D R/W nD R/W |
MantidPlot is a version of QtiPlot customised to present a technique-independent data analysis framework for Neutron and Muon data It is built around C++/Python2 and supports Python plugins and scripting. The new (2019) Mantid v4, or "Workbench", is built for Python3 and uses MatplotLib graphics. |
n/a |
|||||
|
pyFAI |
no |
A Python library, which can use GPU, for high performance azimuthal integration and calibration of data from area detectors. |
Jérôme Kieffer et al |
|||||
|
QtiKWS |
no |
QtiKWS builds on QtiPlot 0.08.9 and provides a variety of reduction and visualization as well as fit1d and fit2 options |
Vitaliy Pipich |
n/a |
||||
|
SAXSView |
1D R/W |
Small suite of applications for reading, converting and displaying 1D and 2D (CBF) SAXS data. |
D Franke |
n/a |
||||
|
ScatterBrain |
no |
IDL runtime application for the reduction and simple analysis of data from the Australian Synchrotron |
Australian Synchrotron. NB: Based on ealier software from beamline ChemMatCARS 15ID at APS |
Webpage. NB: Requires IDL |
W32 |
Linux |
n/a |
OSX |
|
US-SOMO |
no |
Open source, C++/Qt, GUI program. The main program is devoted to the computation of hydrodynamics from atomic-level structures. But the SAS module provides tools for the simulation of I(q) and P(r) curves from structure utilizing a variety of Debye calculators and the fitting of large numbers of curves against experimental data. Includes an advanced module for Gaussian decomposition of SEC-SAXS data. |
E Brookes, B Demeler & M Rocco | Documentation | Windows | Linux | Unix | Mac |
|
XOTOKO |
no |
An antiquated line-mode application for the manipulation of 1D SAXS data. |
P Bendall, M Koch, J Bordas & G Mant | Documentation | n/a | Linux | Solaris | n/a |
|
Application |
canSAS Format |
Description |
Author |
Documentation |
Windows |
Linux |
Unix |
Mac |
|
FISH |
1D R |
A sophisticated, tried & tested, model-fitting program with a Java GUI developed at ISIS. |
n/a |
n/a |
||||
|
IRENA |
1D R |
IGOR script application providing a range of SAS data modelling tools. NB: Requires IGOR. |
J Ilavsky |
Windows | Mac | |||
|
'NIST' |
IGOR script application for the modelling of SANS/USANS data. |
S Kline |
Webpage. NB: Requires IGOR. |
|||||
|
PRINSAS |
no |
Program for the processing and interpretation of small-angle scattering data tailored to the analysis of sedimentary rocks |
A Hinde |
Documentation. NB: Requires Microsoft Excel & Microsoft Access |
ask author | n/a | n/a | n/a |
|
pySAXS |
no |
GUI for SAXS data treatment (absolute scaling, processing of data, etc) and fitting (Open Source) |
Documentation - NB: Requires Python |
W32/W64 |
Linux32/Linux64 |
Unix |
OSX |
|
|
QtiKWS |
no |
QtiKWS builds on QtiPlot 0.08.9 and provides a variety of reduction and visualization as well as fit1d and fit2 options |
Vitaliy Pipich |
windows
|
Linux
|
n/a | OSX
|
|
|
SASView |
1D R/W nD R/W |
A sophisticated model-fitting program built around C++/Python, supporting Python model plugins, and utilising NIST-developed model functions. |
SasView Developers - NB: Originally funded by the DANSE Project |
W32 (versions prior to 4.2) W64 (versions from 4.2) |
||||
|
SASfit |
|
Program fo rthe plotting and analysis of SAS data. |
J Kohlbrecher |
n/a |
||||
|
SCATTER |
|
Sophisticated program for the analysis of 1D & 2D data from ordered systems |
S Forster |
n/a |
n/a |
n/a |
||
|
US-SOMO |
no |
Open source, C++/Qt, GUI program. The main program is devoted to the computation of hydrodynamics from atomic-level structures. But the SAS module provides tools for the simulation of I(q) and P(r) curves from structure utilizing a variety of Debye calculators and the fitting of large numbers of curves against experimental data. Includes an advanced module for Gaussian decomposition of SEC-SAXS data. |
E Brookes, B Demeler & M Rocco |
|||||
|
WillItFit |
C/Python framework for constructing complex models tailored to specific promlems |
M Pedersen |
||||||
|
X+ |
no |
Program for the analysis of solution SAXS data |
U Raviv |
n/a |
n/a |
n/a |
||
|
Application |
canSAS Format |
Description |
Author |
Documentation |
Windows |
Linux |
Unix |
Mac |
|
ATSAS |
|
An extensive and sophisticated program suite for the analysis of small-angle scattering data from biological macromolecules. |
D Svergun |
n/a |
||||
|
AXES |
no |
Online fitting of SAXS data to atomistic models of proteins/RNA/DNA with explicit solvent |
A Grishaev |
Webpage | n/a | n/a | n/a | n/a |
|
BioXTAS RAW |
no |
Open-source, python, multiplatform, GUI-based program focused on easy reduction of 2D data to 1D data and preliminary analysis of 1D data for biological SAS experiments. |
S Skou & J Hopkins |
Website | Windows | Linux | Unix | OSX |
|
CCP-SAS |
no |
Online atomistic simulation of biological macromolecules (and some soft matter systems) - incoporates SCT/SCTPL, US-SOMO & SASSIE |
CCP-SAS |
Webpage | ||||
|
D+ |
no |
Reciprocal grid based algorithm software for modelling the solution scattering from supramolecular structures at different levels of spatial resolution |
U Raviv et al |
W64 | n/a | n/a | n/a | |
|
decodeSAXS |
no |
Deep learning method based on the auto-encoder framework for model reconstruction from small angle scattering data |
H Liu |
|||||
|
DENFERT |
no |
Ab-initio structural modelling of biological macromolecules (ie, similar to DAMMIN in ATSAS) but including the contribution of the inherent hydration layer. |
A Koutsioubas & J Perez |
Webpage | W32 | Ubuntu | n/a | OSX |
|
DENSS |
no |
Calculates 3D particle electron densities directly from 1D solution scattering data |
T Grant |
Webpage | Python | Python | Python | Python |
|
DISORDER |
no |
Simulates the diffraction from fibril assemblies and optimises fibril models with respect to the experimental diffraction data |
A Borovinskiy & Fibernet |
Webpage | W32 | n/a | n/a | n/a |
|
FD2BSL |
no |
Converts ASCII LSQINT (or FDSCALE) output files to BSL/OTOKO format. |
CCP13 |
Documentation | W32 | Linux | Solaris7/Irix6.2/OSF1 | n/a |
|
FDSCALE |
no |
Scales LSQINT intensity output files. |
CCP13 |
Documentation | W32 | Linux | Solaris7/Irix6.2/OSF1 | n/a |
|
FreeSAS |
no |
A Python reimpementation of some ATSAS small angle scattering tools |
G Bonamis & J Kieffer |
Documentation | Python | Python | Python | Python |
|
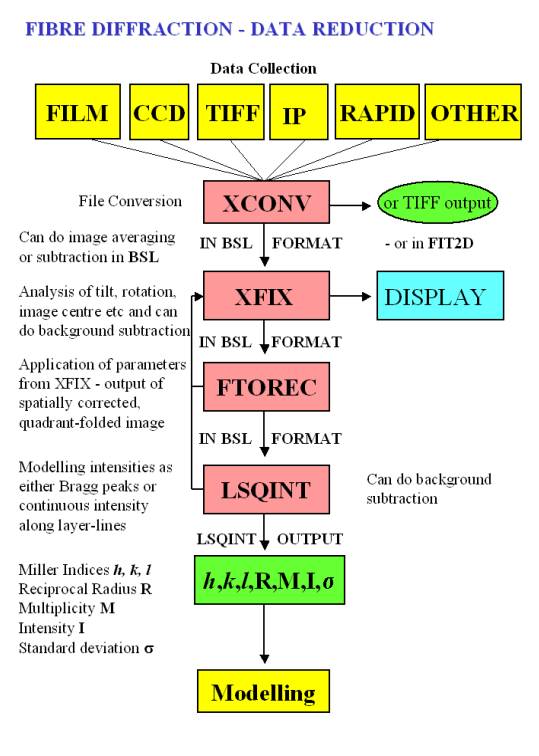
FTOREC |
no |
Transforms all or part of a diffraction image into reciprocal space. NB: Also see LSQINT |
CCP13 |
Documentation | see FibreFix | Linux | Solaris7/Irix6.2/OSF1 | n/a |
|
FibreFix |
no |
A GUI-driven program environment integrating XCONV, XFIX, FTOREC & LSQINT NB: this version requires MS NET Framework 1 which is incompatible with Windows 7 |
CCP13 |
Documentation | W32 | n/a | n/a | n/a |
|
FibreFixWin7 |
no |
NB: use this version for Windows 7 and higher |
A He | - | W32 | n/a | n/a | n/a |
|
FibreFixJava |
no |
NB: this version runs within Java SE | G Rajkumar | - | Java | n/a | n/a | n/a |
|
HELIX |
no |
Simulates the diffraction from helices. |
C Knupp & J Squire | Documentation | W32 | n/a | n/a | n/a |
|
LSQINT |
no |
Transforms all or part of a diffraction image into reciprocal space. NB: Also see FTOREC |
CCP13 |
Documentation | see FibreFix | Linux | Solaris7/Irix6.2/OSF1 | n/a |
|
MusLABEL |
no |
Simulates & labels the diffraction from striated muscle fibres. |
J Squire & C Knupp |
Documentation | W32 | n/a | n/a | n/a |
|
Sample |
no |
Performs Fourier-Bessel smoothing on continuous layer line data. |
CCP13 |
Documentation | W32 | Linux | Solaris7/Irix6.2/OSF1 | n/a |
|
WAXSiS |
no |
Online computation of small- and wide-angle X-ray scattering curves based on explicit-solvent all-atom molecular dynamics simulations. |
Website | |||||
|
WCEN |
no |
A fiber diffraction pattern processing program |
Webpage | W32 | Linux | n/a | OSX | |
|
WinLALS |
no |
Linked-Atom Least-Squares (LALS) program for building molecular and crystal structures of helical polymers and refining them to fit X-ray fiber diffraction data. |
K Okuyama |
n/a |
n/a |
n/a |
||
|
XFIX |
no |
A GUI-driven program for processing fibre diffraction patterns and extracting important parameters. |
CCP13 |
see FibreFix |
n/a |
|||
|
XPLOR/CNS |
no |
A very sophisticated biomacromolecular structure determination program. NB: CNS is a collaborative extension of the original XPLOR. |
A Brünger et al/NIH |
n/a |
n/a |
|||
| Several of the programs in this section originate from CCP13 (Collaborative Computational Project #13). A flowchart of how some of them are integrated into the CCP13 analysis suite may be viewed here. | ||||||||
| Software for Computing Scattering from Coordinate Data | ||||||||
| Application | canSAS Format | Description | Author | Documentation | Windows | Linux | Unix | Mac |
| ATSAS | D Svergun | Programs | W32/W64 | RedHat/Ubuntu/Debian | n/a | OSX | ||
| CCP-SAS | no |
Online atomistic simulation of biological macromolecules (and some soft matter systems) - incoporates SCT/SCTPL, US-SOMO & SASSIE |
CCP-SAS | Webpage | ||||
| Model2SAS | no |
Program to generate small angle scattering curve from 3D models. Models can be STL file and/or math description by .py file. Both module and GUI are available for convenience. |
P Yin | |||||
| SasView |
1D R/W nD R/W |
A sophisticated model-fitting program built around C++/Python, supporting Python model plugins, and utilising NIST-developed model functions. | SasView Developers - NB: Originally funded by the DANSE Project |
W32 (versions prior to 4.2) W64 (versions from 4.2) |
n/a | Debian | OSX | |
| SimSAXSLee | no |
Simulates small & wide-angle scattering, in 1D or 2D, including GISAXS & GIWAXS geometries. Scattering from a single particle or assemblies of particles can be simulated |
B Lee | Website | ||||
| US-SOMO | no |
Open source, C++/Qt, GUI program. The main program is devoted to the computation of hydrodynamics from atomic-level structures. But the SAS module provides tools for the simulation of I(q) and P(r) curves from structure utilizing a variety of Debye calculators and the fitting of large numbers of curves against experimental data. Includes an advanced module for Gaussian decomposition of SEC-SAXS data. |
E Brookes, B Demeler & M Rocco | Documentation | Windows | Linux | Unix | Mac |
|
Application |
canSAS Format |
Description |
Author |
Documentation |
Windows |
Linux |
Unix |
Mac |
|
CORFUNC |
no |
Computes 1D & 3D density correlation functions from SAS data. |
CCP13 |
Overview | n/a | Linux | Solaris7/Irix6.2 | n/a |
|
CORFUNC Java |
no |
Computes 1D & 3D density correlation functions, and adsorbed layer segment density distributions, from SAS data. |
CCP13 & S King |
W32 | n/a | n/a | n/a | |
|
XFIT |
no |
A GUI-driven peak-fitting program. |
CCP13 |
Documentation | n/a | Linux | Solaris7/Irix6.2/OSF1 | n/a |
|
Application |
canSAS Format |
Description |
Author |
Documentation |
Windows |
Linux |
Unix |
Mac |
|
McSAS |
no |
Extracts free-form size distributions |
B Pauw |
Downloads | ||||
|
Application |
canSAS Format |
Description |
Author |
Documentation |
Windows |
Linux |
Unix |
Mac |
|
CELLREF |
no |
Lattice refinement routine. |
K Gardner |
See source | Source | |||
|
DSPACE |
no |
Calculates and sorts d-spacings. |
K Gardner |
See source | W32 | Source | ||
|
EXPAND |
no |
Expands a BSL format data frame. |
D Marvin |
See install | n/a | Linux | n/a | n/a |
|
FIT2D2RKH |
no |
Simple utility to convert FIT2D 1D ASCII format data into ISIS SANS 1D ASCII format data. Useful as a prelude to converting the data into BSL format with XCONV |
S King |
n/a |
n/a |
n/a |
n/a |
|
|
FreeSAS |
no |
A Python reimpementation of some ATSAS small angle scattering tools |
G Bonamis & J Kieffer |
|||||
|
PDH2RKH |
no |
Simple utility to convert SAXSess PDH 1D ASCII format data into ISIS SANS 1D ASCII format data. Useful as a prelude to converting the data into BSL format with XCONV |
S King |
n/a |
n/a |
n/a |
n/a |
|
|
PROP |
Utility that provides an environment for legacy programs ported from Unix/Linux to work comfortably under Windows. provides a means to run the original ILL SANS programs under Windows |
R Ghosh |
Documentation |
n/a |
n/a |
n/a |
||
|
RKH2NIST |
no |
Simple utility to convert ISIS SANS 2D ASCII format data into NIST 2D IGOR format ASCII data. Useful as a means for 2D fitting ISIS data in SansView
|
S King |
n/a |
n/a |
n/a |
n/a |
|
|
XCONV |
no |
Converts several different X-ray detector (and reduced ASCII format ISIS SANS) data formats to BSL format. |
CCP13 |
W32 | Linux | Solaris7/Irix6.2/OSF1 | n/a | |
|
Application |
canSAS Format |
Description |
Author |
Documentation |
Windows |
Linux |
Unix |
Mac |
| Datasqueeze |
yes |
A graphical interface for analyzing data from two-dimensional x-ray diffraction detectors, including small-angle data. (10 day free trial) |
P Heiney | W32/W64 | Linux | Solaris | OSX | |
| DIFFRAC plus |
no |
Bruker | ||||||
| EasySAXS |
no |
PANalytical | ||||||
| SAXSanalysis |
no |
Software for data reduction, handling, and analysis of scattering data | Anton Paar | W32/W64 | n/a | n/a | n/a | |
| SAXSLab |
tbc |
Data collection and reduction package with some limited analysis capability for Rigaku BioSAXS systems. Additional analysis capability is available with an add-on module. | Rigaku | Website | W64 | CentOS 6 | ||
| SAXSquant |
no |
SAXS data collection and basic data handling/analysis (legacy software; see SAXSanalysis) | Anton Paar | W32/W64 | n/a | n/a | n/a | |
|
Software for the Analysis of Grazing Incidence SAS Also see http://gisaxs.com/index.php/Software |
||||||||
| Actionjava | Analysis of SAXS, ASAXS, and GISAXS data using distorted-wave Born approximation. French and English versions. NB: requires Java. | Olivier Lyon | WXP/W7/W8 | Unix | MacOS | |||
| BornAgain | Simulate and fit GISANS and GISAXS data using distorted-wave Born approximation. | Scientific Computing Group at MLZ Garching | W7/W8 | Linux32/Linux64 | OSX | |||
| DPDAK |
Open source tool for (online) analyzing large sequences of small angle scattering data. NB: Has FIT2D and GISAXS plugins. |
DESY/MPIKG | Website | Windows | Linux | |||
| fitGISAXS | Modelling & analysis of GISAXS data using distorted-wave Born approximation. NB: requires IGOR. | D Babonneau | Website | |||||
| HiPGISAXS | Massively-parallel HPC code for simulating GISAXS | HiPGISAXS Team at LBL | Documentation | Linux64 | ||||
| IsGISAXS | Simulates and analyses Grazing Incidence Small Angle X-Ray Scattering (GISAXS) from nanostructures. | R Lazzari | Website | W32 | n/a | n/a | n/a | |
| SimSAXSLee | Simulates small & wide-angle scattering, in 1D or 2D, including GISAXS & GIWAXS geometries. Scattering from a single particle or assemblies of particles can be simulated. | B Lee | Website | |||||
| Other SAS Software | ||||||||
|
List of SANS Analysis Programs installed at the ILL (R Ghosh) |
||||||||
| The ALS SAXS Toolbox | ||||||||
| The ESRF SAXS Program Package (P Boesecke) | ||||||||
| Source Code Repositories | ||||||||
| scattering-central | ||||||||
| Software for the Analysis of Reflectometry Data | ||||||||
| Compilation by Adrian Rennie | ||||||||